There are increased demands for high sensitivity, selectivity, and rapidity sensing devices to meet the needs of in-clinical diagnostics and epidemic prevention. Homogeneous assays for target molecules with fluorogenic probes have attracted intense attentions due to their operational convenience and ease of automation. Single-layer 2D nanosheets (NSs) have demonstrated to be a class of efficient sensing platform for detecting deoxyribonucleic acid (DNA) and small molecules, attributing to their selective adsorption and fluorescence-quenching abilities toward dyelabeled single-stranded DNA (ssDNA).
Graphene oxide (GO), a water-soluble derivative of graphene (GR), was the earliest reported 2D materials to possess fluorescence quenching properties. Due to the π-π stacking interactions between graphene and nucleic acid, ssDNAs can be strongly adsorbed on GO, resulting in a substantial fluorescence quench of the organic dyes labeled on ssDNAs through the fluorescence resonance energy transfer between the hexagonal cells of graphene and the dye molecular. However, double-stranded DNAs (dsDNA) cannot quench the fluorescence because of their negatively charged phosphate backbone will shield the nucleobases to reduce the interactive force between the dsDNA and GO.
Being a new 2D carbon materials, the unique structural characteristics of graphdiyne (GD) makes it very promising for the biosensing applications. Recently, researchers’ density functional theory calculation results suggest that the dye molecular absorption on GD is stronger than that on GR. Moreover, the GD NSs exhibits a superior electrons capturing ability than that of GR. The electrochemical lithium-intercalation method was applied to prepare the few-layered GD NSs. Fig 1 shows the TEM images of the GD NSs with and without lithium-intercalation treatment. The AFM data reveal that the thickness of the as-prepared GD is about 1.1 nm, and such a thickness is comparable to those common 2D MoS2 and GO. For the first time, few-layer GD NSs have been demonstrated to possess high fluorescence quenching abilities and different affinities toward the ssDNA versus dsDNA. Such superior properties of GD NSs can be advantageously used to develop new biosensing principles for multiplexed real-time fluorescent detection of DNA in a highly sensitive manner with a limit of detection as low as 25 × 10-12 M (Fig 2). Importantly, comparing with GO and MoS2 nanomaterials-based sensors, the GD NSs-based biosensor exhibits high sensitivity and short detection time for detection of multiplexed DNA. In addition, the assay can be carried out in homogeneous liquid phase, making it perfectly suits the in situ detection applications. The efficient GD nanoquenchers can be readily synthesized in large-area which can be loaded with different dye-labeled ssDNA make the material have the advantages for analysis of multiplexed DNA. We expect that the GD NSs nanoprobes demonstrated in this work for facile, rapid, and cost effective multiplexed detection of biological molecules would pave a way for widespread biological analysis using 2D nanomaterials-based biosensing systems.
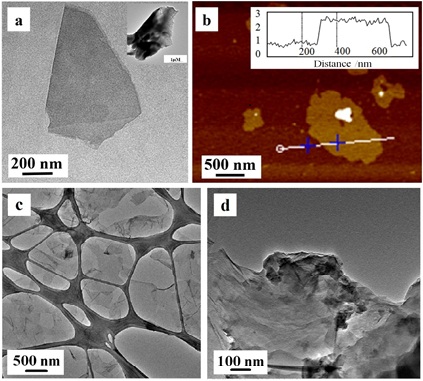
Fig 1. a) TEM image of a typical GD nanosheet after lithium-intercalation treatment (inset: GD
nanosheet without lithium-intercalation treatment); b) AFM image of GD nanosheets, deposited on Si/SiO2 substrate (inset: Height profile of the dotted line, showing thickness of ~1.1 nm); c) TEM image of MoS2; d) TEM image of GO.
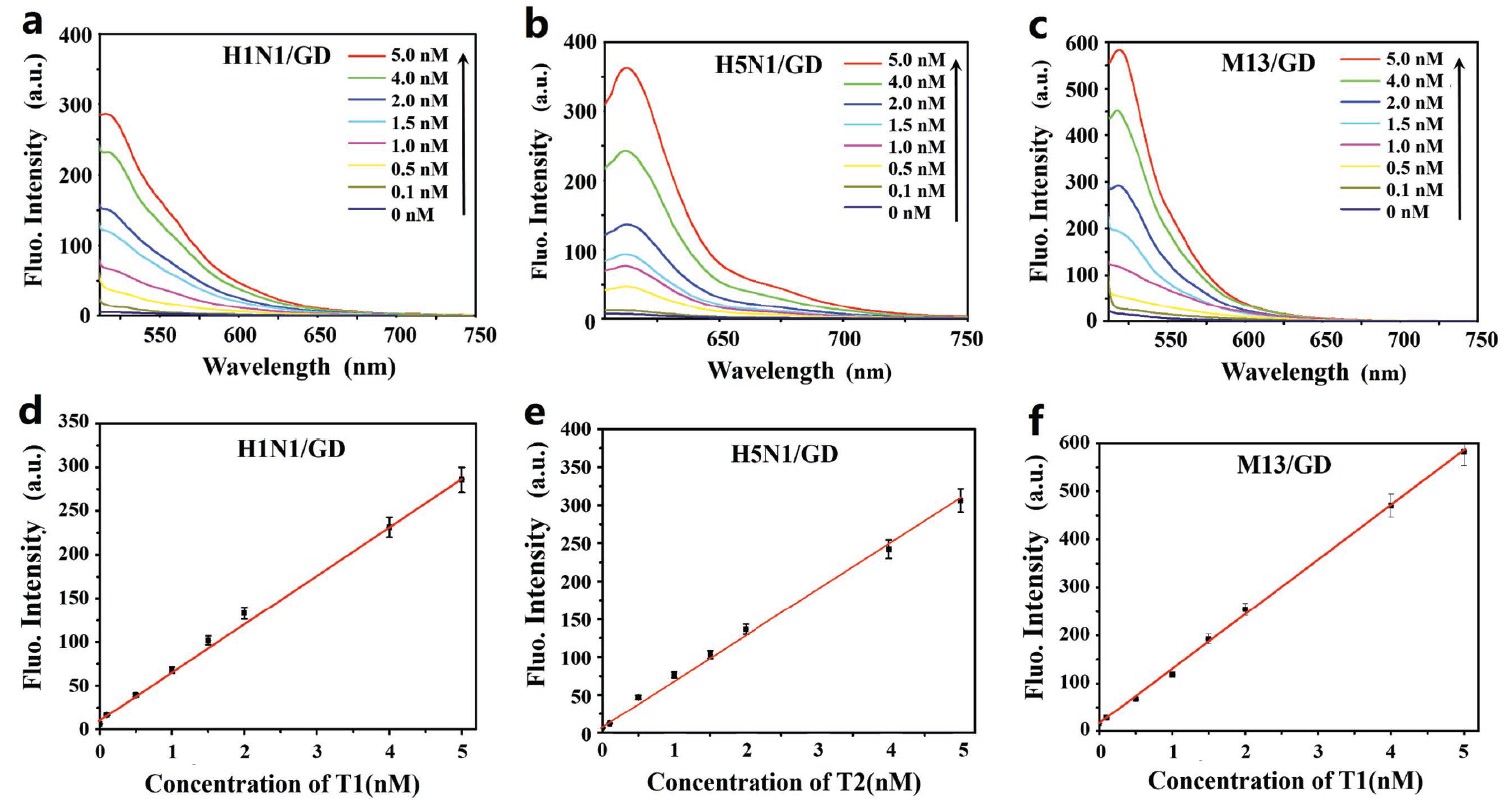
Fig 2. a-c) Fluorescence spectra of the dye-labeled ssDNA (P1:H1N1-FAM, P2: H5N1-Texas Red, P3:M13-FAM) in the presence of various concentrations of target DNA (T1, T2, and T3, 0-5 × 10?9 M); d–f) Calibration curve for target DNA detection. GD was 20.0 μg mL-1. With the excitation/emission wavelengths of a,c) 494 nm/516 nm and b) 595 nm/612 nm, respectively.
The research team includes a group led by Prof. WANG Dan from the Institute of Process Engineering (IPE) of the Chinese Academy of Sciences (CAS), a group led by YU Ranbo from University of Science & Technology Beijing, a group led by HUANG Ling from Nanjing Tech University, a group led by WANG Lianhui from Nanjing University of Posts and Telecommunications and a group led by ZHAO Huijun from Griffith University. The research was supported by the National Science Fund for Distinguished Young Scholars (No. 21325105), National Natural Science Foundation of China (Nos. 21590795, 51672276, 21671016, 51372245, 51541206), CAS Interdisciplinary Innovation Team, the Foundation for State Key Laboratory of Biochemical Engineering, and et al.
Their work entitled “Few-layer graphdiyne nanosheets applied for multiplexed real-time DNA detection” has been published in Adv. Mater (2017, 29, 1606755).
http://onlinelibrary.wiley.com/doi/10.1002/adma.201606755/full
Media Contact:
Ms. YUAN Pei
International Cooperation Office, Institute of Process Engineering, Chinese Academy of Sciences, Beijing 100190, P. R. China.
E-mail: pyuan@ipe.ac.cn
Tel:86-10-82544882
 Search
Search




 京公网安备110402500047号
京公网安备110402500047号